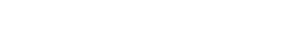The psycopg3 project is currently under active development.
Sponsoring this project will help achieve swift completion and ensure the maintenance of psycopg2, psycopg3 and other related projects.
If you use Python and PostgreSQL, and you would like to support the creation of the most advanced adapter between the two systems, please consider becoming a sponsor. Thank you!
I have been cheerfully hacking on psycopg3 features for a while and reached the symbolic but relevant milestone of DBAPI 2.0 compatibility while using the psycopg2 test suite as a guideline to add features progressively.
Then I thought about checking its speed... It wasn't a happy day.
A few years ago, a new PostgreSQL driver was released: asyncpg. It is designed with speed in mind, at the expense of much else. It parses the PostgreSQL frontend/backend protocol using its own implementation (instead of using libpq, the PostgreSQL client library) and is designed to be pretty much streamlined within the flow of the line protocol rather than have a developer-friendly or recognisable interface: it doesn't follow a DBAPI design, it uses PostgreSQL native placeholders for queries, i.e. where someting = $1, no positional or named ones (the familiar %s and %(thing)s). It is definitely a fast library, but much like a Formula 1 car, it's not for everyone to drive. Dropping it in your existing project means having to rewrite all your queries and all the Python code interacting with the database: no Django, no SQLAlchemy, and of course it's only asynchronous, so not the natural choice if your program uses a different concurrency model or needs none at all.
Anyway I thought I'd use the toolbench they wrote to compare speeds between psycopg2 and asyncpg. So how bad was my day?
| Driver | Queries |
|---|---|
| asyncpg | 15263 |
| psycopg2 + aiopg | 8871 |
| psycopg3 async | 426 |
It was this bad.
This is the number of queries that are run in 10 seconds. The numbers themselves are to be taken with a fistful of salt. Essentially they are obtaining connecting to a database on localhost so they are more CPU-bound than the environment a database driver usually lives in. They come from one specific query (the first in their toolbench) so nothing statistically sound, but still a pretty depressing picture.
Back then, the implementation of psycopg3 was pure Python, using ctypes to call into the libpq shared library. The plan to write an optimised C implementation was already there, but this made me understand how badly needed this is.
Profiling
The first step to climb such a mountain is to look at profiling information, which can show function by function where time is spent.
The graph is generated by gprof2dot using something like:
python -m cProfile -o OUT.pstat script.py gprof2dot -f pstats OUT.pstat | dot -Tsvg > OUT.svg
You can see more details by clicking on the above graph. The left branch is initialisation faff. The interesting part is on the right, following the path of orange, yellow and green. This shows the functions calls where the program spends most of its time. get_value is a pretty hot one, taking up about the 50% of the run time. It gets called very often, once per field of every record, or 1.6M times in this graph. At this point, it looked like this:
def get_value(self, row_number: int, column_number: int -> Optional[bytes]: length = impl.PQgetlength( self.pgresult_ptr, row_number, column_number ) if length: v = impl.PQgetvalue(self.pgresult_ptr, row_number, column_number) return string_at(v, length) else: if impl.PQgetisnull(self.pgresult_ptr, row_number, column_number): return None else: return b""
This function returns the content in bytes of a field. If it's 0-length, it checks if it's NULL. The impl functions are ctypes calls to the libpq. This made me think that the overhead of ctypes calls is not so tiny after all.
The cffi attempt
One thing I thought about doing was also to provide a cffi wrapper for the libpq. cffi is a more popular choice than ctypes, especially thanks to its good integration with PyPy. ctypes has the advantage of being packaged in the standard library, so not requiring an external dependency. However, unlike ctypes, cffi still requires a compiler to create a more efficient wrapper.
Porting the libpq wrapper to cffi was relatively straightforward and the number of queries that ran was more than double:
| Implementation | Commit | Queries |
|---|---|---|
| ctypes | 7b3b7f9 | 426 |
| cffi | dfe2893 | 902 |
Profiling showed that the time spent in get_value was down from 50% to 35%, suggesting that the cffi overhead is lower than ctypes. However, building a cffi wrapper already requires the presence of a C compiler, the libpq-dev package and finding the dreaded pg_config executable: the same requirements a C extension implementation would have, but with less possibilities for optimisation. For this reason, cffi will probably remain as a thing on the side (maybe if the PyPy folks would be interested in it). This code is now parked in its own branch.
The Cython attempt
The next and more reasonable way to improve performance is to create a C extension. As highlighted in Thinking psycopg3, the idea was already in place, allowing the user to choose between the simple to install option or the fast option. A Python C extension can be written in pure C, as psycopg2 currently is, but there are other languages helping to handle some of the tedious aspects, such as the boilerplate required to create the objects, and the reference count. The most current of such languages is Cython, an evolution of Pyrex, which I had once used a ridiculous number of years ago.
The first step was a straightforward porting of the psycogp3.pq module: that alone showed a promising class of results:
| Implementation | Commit | Queries |
|---|---|---|
| ctypes | 7b3b7f9 | 426 |
| cffi | dfe2893 | 902 |
| Cython pq | 6c6cbe3 | 1618 |
Speeding up adaptation
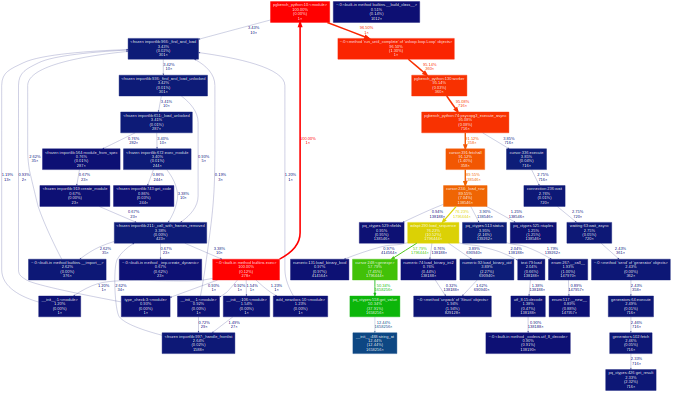
New profiling showed that the time spent in get_value() was now reduced to a paltry 6% of the whole runtime: this means that that the hot path of the program had moved to a different place.
The entire adaptation of the returned dataset, visible in green, takes 60% of the runtime, so is a likely next candidate for optimisation. The <genexpr> box is shown having a large number of called functions: that function is the dispatcher choosing a different adapter function for each PostgreSQL data type, which were still written in Python.
There is probably some value in creating analogous conversion functions in C, but it wouldn't be the best way to optimise the job. All the loader functions have the same signature, for instance: the loader for the binary representation of a 32 bits integer is:
_int4_struct = struct.Struct("!i") def load_int4_binary(data: bytes) -> int: return _int4_struct.unpack(data)[0]
In order to feed data to this function it is necessary to:
- get the pointer and length to the returned data from the PGresult struct (calls to PQgetvalue and PQgetlength, which are lightweight);
- convert it to a Python bytes object, meaning a memcpy and a Python object allocation;
- calling the adaptation function (Python or C).
If we decide not to leave the C layer in the adaptation phase then a better signature can be used:
cdef object load_int4_binary_c(const char *data, size_t length, void *context): return PyLong_FromLong(<int32_t>be32toh((<uint32_t *>data)[0]))
This allows for reaching of the loader functions without the need of an extra copy of the data received from the network and without creating an intermediate Python object. However, because the C functions are not accessible from Python, it poses the question of how to customise the adapters mapping, in case someone wanted to replace a builtin one (a typical case involves wanting to convert numeric into Python float instead of Decimal). For now, the mechanism works like this:
At import time, the Python adapters are associated to data types OIDs; the current mechanism is through a decorator:
# in psycopg3/types/numeric.py @Loader.binary(builtins["int4"].oid) def load_int4_binary(data: bytes) -> int: return _int4_struct.unpack(data)[0]
If the C extension module is to be used, a C function exposed to Python is called, register_builtin_c_loaders().
# in psycopg3/__init__.py # register default adapters from . import types # Override adapters with fast version if available if pq.__impl__ == "c": from ._psycopg3 import register_builtin_c_loaders register_builtin_c_loaders()
This in turns calls a registration function for all the loaders, for which there is a C implementation:
# in psycopg3/types/numeric.pyx from psycopg3.types import numeric register_c_loader(numeric.load_int4_binary, load_int4_binary_c)
When the result of a query is returned to the client, the OIDs of the columns are used to select a Python loader. In Python, the loaders are put into a list, having as many items as the columns number. In Cython, they are put into a C array of structures holding both a Python function and optionally a C function as members. If a Python loader has an optimised version registered then that function is added to the array of loaders.
Type OID Python loader C loader text 25 load_text_binary load_text_binary_c int4 23 load_int4_binary load_int4_binary_c custom ?? load_custom_type NULL When the data is converted to Python, the corresponding loader is used for each column:
- if it has a C loader, the value and lengths are read from the result and passed to the function;
- if it doesn't have one, a bytes object is created and passed to the Python function.
This mechanism allows for the overriding of the mapping from OID to types in Python, which is then duplicated in C. It has the disadvantage that if someone wanted to derive an adapter for a custom data type (e.g. calling the text adapter and then parsing the result) then the Python adapter would be used. There is probably possibility to improve this design, but it is a good starting point for now.
It might be possible to avoid memcpy by creating a memory view instead of a bytes array, but I haven't quite figured out how to do it in ctypes yet.
Numbers, now, start to appear more promising:
| Implementation | Commit | Queries |
|---|---|---|
| ctypes | 7b3b7f9 | 426 |
| cffi | dfe2893 | 902 |
| Cython pq | 6c6cbe3 | 1618 |
| Cython adapt | b28b389 | 7511 |
Profiling against psycopg2
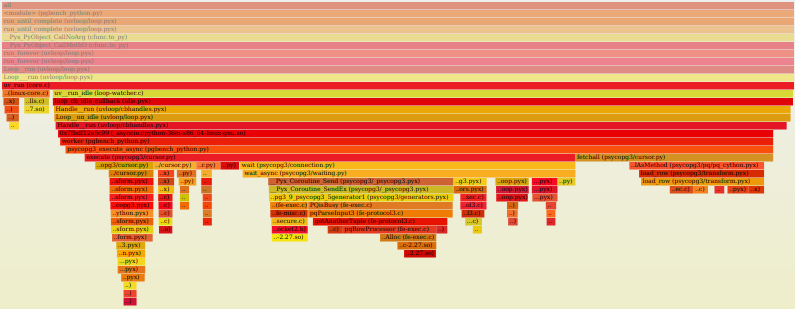
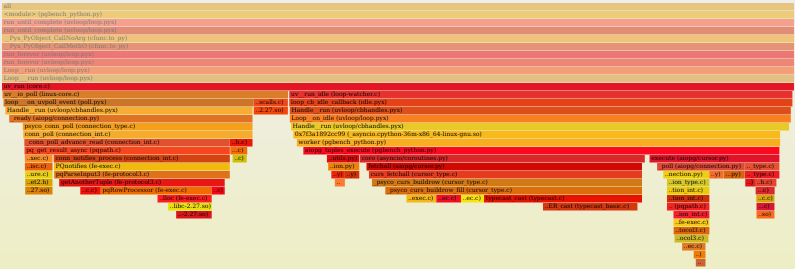
I was having concerns for a while that there was something stupidly wrong in my psycopg3 code, especially around the networking area, for instance introducing delays waiting the wrong way or at the wrong moment. asyncpg is too radically different to provide a direct comparison, but a comparison with psycopg2 would have been interesting. Unfortunately, because psycopg2 is written in pure C code, it doesn't allow inspection using the Python profiler (Cython extensions are C too, but they can create explicit hooks for that). But where a tracing profiler cannot trace, a sampling profiler can sample: py-spy is such a profiler. The principle is to interrupt a process several times per second to look at its call stack and infer what it is doing. Provided enough debug information is compiled in the C library, it is possible to obtain clear information about calls into C code too. And of course, the most important feature good software should always have: the production of pretty graphs:
This comparison shows that the time around the libpq is roughly comparable (psycopg2 calls the chubby pqParseInput3 as a consequence of calling PQnotifies; for psycopg3, it happens in PQisBusy because it currently lacks a notifications handler). There was actually a bug in the fetching loop, but because of that bug, the code was actually running somewhat faster (it would have likely hurt concurrent performance with blocking though).
Current state, future improvements
The diagrams above suggest that there is still room for improvement: fetchall() seems to have some overhead, which can probably be reduced by introducing a "fetchsome" function in C. There is a lot of overhead around waiting, especially in the process of registering/unregistering listeners: the best gains seem achievable here, bypassing some of the asyncio machinery and creating a better integration with the underlying reactor.
Below are the bottom line figures, as of the current master (b662c9d). The pure Python results have improved too, thanks to the Python code tweaking since the first test:
| Driver | Queries |
|---|---|
| asyncpg | 15263 |
| psycopg2 + aiopg | 8871 |
| psycopg3 async Python | 517 |
| psycopg3 async C | 8424 |
Again, this is just a rough indication: the data types in the query and different concurrency patterns create different pictures. For instance, psycopg3 is already faster than psycopg2 in benchmarks involving bytea, thanks to the use of the binary protocol:
| Driver | Queries |
|---|---|
| asyncpg | 41330 |
| psycopg2 + aiopg | 12396 |
| psycopg3 async C | 19164 |
I think for the moment, the search for speed can be concluded and it is time to go back to progressing with the features. I doubt that asyncpg's stellar performances can be passed, but I don't want performance to be the sole goal: I am fine with psycopg3 being somewhat slower than asyncpg, if it has the advantage to be easier to use and to behave pretty much as a drop-in replacement for psycopg2. In my view, a driver with a lower adoption barrier would be more directly beneficial for everyone wanting to use it, either in existing frameworks or on its own. By the time the project will be released, I am confident that its performance will consistently pass the psycopg2 level.